러닝스푼즈 수업 정리
< 이전 글 >
https://silvercoding.tistory.com/62
[boston 데이터 분석] 1. 차원축소 (PCA) 파이썬 예제
러닝스푼즈 수업 정리 라이브러리 & 데이터 불러오기 - 라이브러리 불러오기 import pandas as pd import matplotlib.pyplot as plt import seaborn as sns - 데이터 불러오기 data = pd.read_csv('./data/bosto..
silvercoding.tistory.com
라이브러리 & 데이터 불러오기
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as snsdata = pd.read_csv('./data/boston.csv')
data.head()
군집화 Clustering
del data['chas']연속형 데이터만 다루기 위하여 범주형 데이터는 제거한다.
medv = data['medv']
del data['medv']타겟변수를 복사해 놓고, 타겟변수 컬럼을 지워준다. ( pca를 위하여 )
차원 축소 (PCA) : 12차원 -> 2차원
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA차원 축소에 필요한 라이브러리를 import 해준다.
- 정규화
scaler = StandardScaler()정규화 객체를 생성한다.
# 데이터 학습
scaler.fit(data)
# 변환
scaler_data = scaler.transform(data)data 전체를 정규화하여 scaler_data에 저장해 준다.
- PCA
pca = PCA(n_components = 2)PCA 객체를 생성한다. 2차원 시각화를 위하여 변수는 2개로 설정한다.
pca.fit(scaler_data)pca로 scaler_data를 학습시킨다.
data2 = pd.DataFrame(data = pca.transform(scaler_data), columns=['pc1', 'pc2'])pca로 변환한 데이터를 데이터 프레임으로 data2에 저장한다.
data2.head()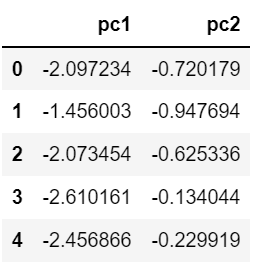
군집의 개수 정하기 - Elbow Point 지정
from sklearn.cluster import KMeansKMeans(n_cluster = k)
- k개의 군집화를 하겠다는 객체 생성
Kmeans.fit()
- 학습시키기
KMeans.inertia_
- 학습된 KMeans의 응집도를 확인
- 응집도란 각 데이터로부터 자신이 속한 군집의 중심까지의 거리를 의미
- 즉, 낮을수록 군집화가 더 잘되어있음.
KMeans.predict(data)
- 학습된 데이터를 바탕으로 데이터를 변환시켜줌
x = [] # k 가 몇개인지
y = [] # 응집도가 몇인지
for k in range(1, 30):
kmeans = KMeans(n_clusters = k)
kmeans.fit(data2)
x.append(k)
y.append(kmeans.inertia_)1부터 30까지 군집화를 해보고, 가장 적절한 응집도의 군집개수를 정해주기 위하여 그래프를 그려본다.
plt.plot(x, y)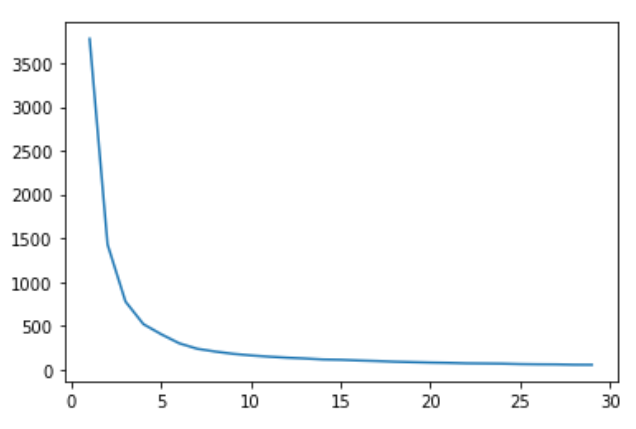
군집의 개수 별 응집도 그래프를 그려보니 3~5개 정도가 적당할 것 같다. Elbow Point를 4로 지정하고 군집화를 해보도록 한다.
군집화
kmeans = KMeans(n_clusters=4)군집의 개수를 4로 설정하여 객체를 생성한다.
kmeans.fit(data2)
위에서 생성해 놓은 data2를 학습한다.
data2['labels'] = kmeans.predict(data2)각각의 예측된 군집 종류를 labels 컬럼에 넣어준다.
data2.head()
lebels가 1이라는 것은 해당 데이터가 1번 군집에 포함되었다는 의미이다.
sns.scatterplot(x='pc1', y='pc2', hue='labels', data=data2)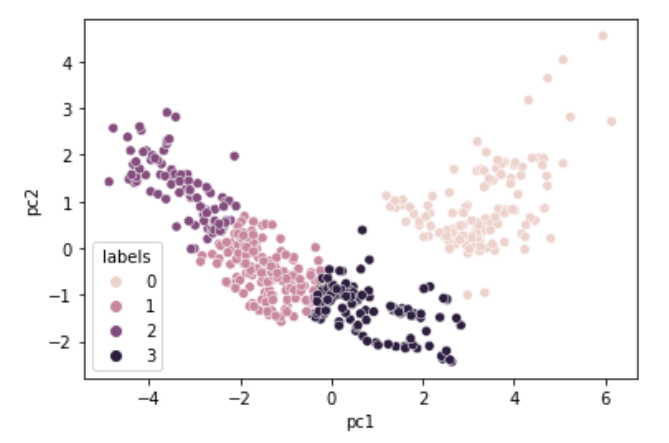
위와 같이 군집이 형성되었음을 확인할 수 있다.
결과 해석
- 어떤 그룹의 집 값이 가장 높을까 ? : 평균으로 비교
data2['medv'] = medv각 그룹의 집값 평균을 구하기 위해 처음에 저장해 주었던 medv 컬럼을 data2의 medv 컬럼으로 추가해 준다.
data2.head()
data2[data2['labels']==0]['medv'].mean()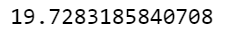
0번 군집의 집값의 평균은 다음과 같이 구한다. 이를 활용하여 모든 군집의 집값의 평균을 그래프로 그려보도록 한다.
medv_list = []
for i in range(4):
medv_avg = data2[data2['labels']==i]['medv'].mean()
medv_list.append(medv_avg)sns.barplot(x=['group_0', 'group_1', 'group_2', 'group_3'], y=medv_list)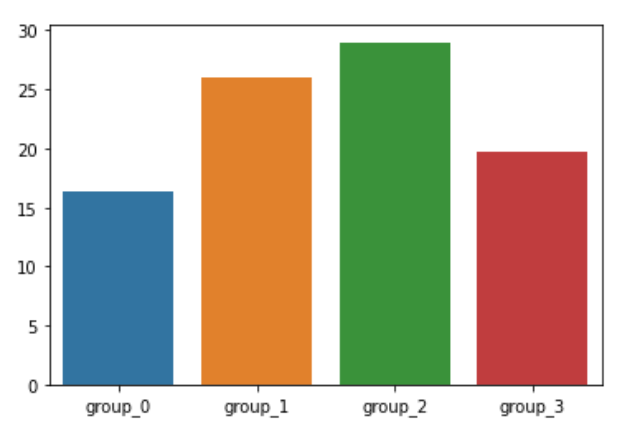
집값의 평균 최상위 그룹 : group_2
집값의 평균 최하위 그룹 : group_0
---> 최상위 그룹과 최하위 그룹을 비교하여 집값의 평균이 높거나 낮은 이유에 대하여 확인해 본다.
* 원본 데이터 사용하여 원인 분석해보기
data['labels'] = data2['labels']원본데이터에 그룹 labels를 추가해 준다.
group = data[(data['labels']==0) | (data['labels']==2)]그룹0, 그룹2 만 선택하여 group 변수에 저장한다.
group = group.groupby('labels').mean().reset_index()gropuby를 사용하여 labels 컬럼 별로 모든 컬럼의 평균값을 구하고, groupby로 인하여 인덱스가 되었던 labels를 reset_index()를 이용하여 다시 컬럼으로 변경해 준다.
group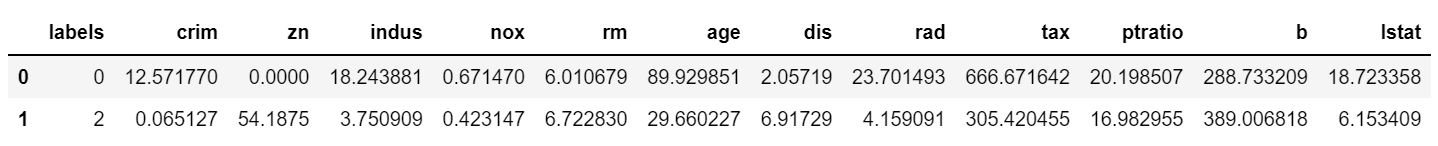
각 그룹별 평균이 구해진 것을 볼 수 있다. 이를 시각화 하여 비교해 보도록 한다.
결과 해석 - 시각화
column = group.columns
fig, ax = plt.subplots(2, 6, figsize=(30, 13))
for i in range(12):
sns.barplot('labels', column[i+1], data=group, ax=ax[i//6, i%6])
두개의 막대가 그려져 있는 막대그래프로, 왼쪽이 높으면 집값이 낮은 이유의 근거가, 오른쪽이 높으면 집값이 높은 이유의 근거가 된다고 해석한다.
결론
- (0,0) 위치의 그래프를 보면 crim( 범죄율 )이 0번 그룹에서 월등히 높다. 이는 범죄율이 높을수록 집값이 낮다고 해석할 수 있다.
- (0, 1) 위치의 그래프에서는 zn( 25,000 평방비트를 초과하는 거주지역 비율 ) 이 높을수록 집값이 높다고 해석된다.
- 그다음으로 차이가 적어 보이는 것은 (1, 1) 위치의 그래프이다. rad( 방사형 고속도로까지의 거리 ) 가 높을수록 ( 방사형 고속도로까지의 거리가 멀수록 ) 집값이 낮다는 것을 알 수 있다.
이와 같이 여러 해석을 해볼 수 있다.
'데이터 분석 이론 > 머신러닝' 카테고리의 다른 글
| [Bank Marketing데이터 분석] 2. python 부스팅 Boosting, XGBoost 사용 (0) | 2021.08.23 |
|---|---|
| [Bank Marketing데이터 분석] 1. python 배깅 , 랜덤포레스트 bagging, randomforest (0) | 2021.08.23 |
| [IRIS 데이터 분석] 2. Python Decision Tree ( 의사 결정 나무 ) (0) | 2021.08.20 |
| [IRIS 데이터 분석] 1. Python KNN 분류 (0) | 2021.08.20 |
| [boston 데이터 분석] 1. 차원축소 (PCA) 파이썬 예제 (0) | 2021.08.18 |